ICCV 2021 Workshop: MIA-COV19D
The Workshop “AI-enabled Medical Image Analysis Workshop and Covid-19 Diagnosis Competition (MIA-COV19D)” will be held in conjunction with International Conference on Computer Vision (ICCV) 2021 in Montreal, Canada, October 11- 17, 2021.
For any requests or enquiries, please contact: stefanos@cs.ntua.gr
Organizers
- Stefanos Kollias (National Technical University Athens, Institute Computer Communication Systems)
- Xujiong Ye ( University of Lincoln ) xye@lincoln.ac.uk
- Luc Bidaut ( University of Lincoln ) lbidaut@lincoln.ac.uk
- Francesco Rundo ( STMicroelectronics ADG—Central R&D ) francesco.rundo@st.com
- Dimitrios Kollias ( University of Greenwich ) d.kollias@qmul.ac.uk
- Giuseppe Banna ( Portsmouth Hospitals Univ. NHS Trust ) giuseppe.banna@nhs.net
Data Chairs
- Anastasios Arsenos ( National Technical University of Athens )
- Manos Seferis ( National Technical University of Athens )
- James Wingate ( University of Lincoln )
Description
There are two activities in the ICCV 2021 AI-enabled MIA-COV19D Workshop:
A. AI-enabled Medical Image Analysis (MIA) Workshop
A1. Scope
AI-enabled Medical Image Analysis (MIA) Workshop is devoted to medical image analysis, with emphasis on radiological quantitative image analysis for diagnosis of diseases. Our focus is placed on Artificial Intelligence (AI), Machine and Deep Learning (ML, DL) approaches that target effective and adaptive diagnosis; we also have a particular interest in approaches that enforce trustworthiness and automatically generate explanations, or justifications of the decision making process.
A2. Call for Participation
Technologies and topics to be addressed include, but are not limited to:
- 2D/3D-CNN, CNN-RNN, GANs and ML models in medical image diagnosis
- Sensing “salient features” of Deep Neural Networks (DNN) and AI models related to decision-making, in spatial (images), temporal (video) and volumetric (3-D) data
- Optimal visualization of salient features and areas in the input data
- Low/Middle/High-level feature extraction, analysis and interpretation
- Exploration of how predictions in data streams are correlated and explain each other (multimodal data)
- Joint optimization of positive and negative saliencies
- Global and local models for prediction, or classification
- Attention and self-attention mechanisms in DNNs/AI approaches
- Explainable AI based on anchors, or attention
- Interpretability at training time through adversarial regularization
- Learning new data (from multiple sources) by leveraging knowledge already extracted and codified, through domain adaptation
- Uncertainty estimation and quantification, self training
- Zero/one shot learning, avoidance of catastrophic forgetting
- Evaluation of explanations generated by ML/DL/AI models in medical imaging
Original high-quality contributions will be targeted in a variety of contexts: in enforcing shared patterns to emerge directly from data; in supporting automated segmentation of CT data; in using visual attention both to improve classification accuracy and to provide interpretability of model outputs; in highlighting the areas in the medical imagery responsible for the provided decisions; in supporting medical staff in their analysis.
A3. Important Dates
For the papers to be submitted to the Workshop (not including the Competition) the timeline will be as follows:
- July 23, 2021: Paper submission deadline; Start of review period NEW DATE
- August 6, 2021: End of review period
- August 10, 2021: Review decisions sent to authors; Notification of acceptance
- August 17, 2021: Camera ready version deadline
A4. Submission Information (UPDATED)
The peer review process for the papers will be double blind, including at least two distinct reviews per submission, and will follow ICCV standards and policies. The accepted papers will be part of ICCV 2021 conference proceedings. All paper submissions must adhere to the ICCV 2021 paper submission style, format, and length restrictions. Please have a look here.
The submission process will be handled through CMT.
A5. Keynote Speakers
- Sebastiano Battiato, Professor, Department of Mathematics & Informatics, University of Catania, “COVID-19 Diagnosis by IA-based imaging technologies”.
- Massimo Villari, Professor, Future Computing Research laboratory, University of Messina, “Neuro-Crying Analysis by IA-based imaging: children crying with neuro disorder”.
- Dimitrios Nikitopoulos, Ph.D, Greek National Infrastructures for Research & Technology, “Advanced Services to Hospital Units in the Cloud: HPC & COVID-19 Diagnosis”.
A6. Workshop Agenda
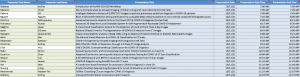
The workshop will take place virtually on October 11, 2021 from 07:00 (am) – 12:00 (pm) EDT (Montreal) time. The workshop agenda can be seen below:
B. COV19D Competition
B1. Scope
COV19D Competition is based on a database of chest CT scan series that is manually annotated with respect to Covid-19/non-Covid-19 diagnosis. The database is split into training, validation and test partitions. The training and validation sets along with their corresponding annotations will be provided to the Competition participating teams to develop AI/ML/DL models for Covid-19 and non-Covid-19 prediction. Performance of the different approaches will be evaluated on the test set.
B2. Call for Participation
The participating teams will submit their Covid-19 predictions for the test dataset. The winner of the Competition will be selected based on the achieved performance, in terms of the ‘macro’ F1 score, on the test set.
There will be one Competition winning team. The top-3 performing teams are expected to contribute a paper describing their approach, methodology and results to our MIA Workshop. All other teams will be able to submit a paper describing their solutions and final results to the MIA Workshop. The accepted papers will be part of the ICCV 2021 proceedings.
Regarding the database: It consists of about 5,000 CT scan series. Each chest CT scan series consists of a sequence of 2-D CT slices, the number of which is between 50 and 700. The research groups will be able to use any publicly available datasets as long as they report it in their submitted final paper. We will provide a white paper describing the Competition, the database used, as well as a baseline model and its results for comparison purposes.
In order to participate in the Competition, teams will need to register. At the end of the Competition, each team will have to send us: i) their predictions on the test set, ii) a link to a Github repository where their solution/source code will be stored, and iii) a link to an ArXiv paper with 2-6 pages describing their proposed methodology, data used and results. After that, the winner of the Competition will be announced and a leaderboard will be published. The winner and the two runner-ups in the Competition will be asked to also share their trained models so as to check out the validity of the approach.
B3. Important Dates
The timeline for the competition will be as follows:
- May 3, 2021: Opening of the Competition; start of registration of research groups; provision of training and validation datasets, of respective annotations and of the baseline approach
- July 10, 2021: Final submission deadline (Results, Code and ArXiv paper) NEW DATE
- July 11, 2021: Winners announcement NEW DATE
- July 23, 2021: Final paper submission deadline NEW DATE
- August 6, 2021: End of review period
- August 10, 2021: Review decisions sent to authors; Notification of acceptance
- August 17, 2021: Camera ready version deadline
B4. Participation Information
To participate, you need to register your team.
For this, please send us an email to: d.kollias@qmul.ac.uk
with the title “ICCV COV19D Competition: Team Registration”.
In this email include the following information:
Team Name
Team Members
Affiliation
There is no maximum number of participants in each team.
As a reply, you will receive the participation information and then access to the dataset’s images and annotations.
B5. Final Paper Submission Information
The peer review process for the papers will be double blind, including at least two distinct reviews per submission, and will follow ICCV standards and policies. The accepted papers will be part of ICCV 2021 conference proceedings. All paper submissions must adhere to the ICCV 2021 paper submission style, format, and length restrictions. Please have a look here.
The submission process will be handled through CMT.
B6. Competition Results (NEW!)
35 Teams participated in the COV19D Competition.
18 Teams submitted their results.
12 Teams scored higher than the baseline and made valid submissions; their results are shown in the leaderboard below.
The winner of the Competition is FDVTS_COVID consisting of: Junlin Hou, Jilan Xu, Rui Feng, and Yuejie Zhan (Fudan University, China).
The runner-up (with a slight difference from the winning team) is SenticLab.UAIC consisting of: Mihaela Breaban, Cosmin Moisii, Radu Miron, Vlad Barbu and Sergiu Dinu (SenticLab and “Alexandru Ioan Cuza” University of Iasi, Romania).
The team that ranked third is ACVLab consisting of: Chih-Chung Hsu, Kuan-Lin Chen, Chieh Lee and Mei-Hsuan Wu (National Cheng Kung University, Taiwan)
The leaderboard can be found here: iccv_cov19d_leaderboard
Congratulations to you all, winning and non-winning teams! Thank you very much for participating in our Competition.
All teams are invited to submit their methodologies-papers (please see B5 above). All accepted papers will be part of the ICCV 2021 proceedings.
We are looking forward to receiving your submissions!
References
- D. Kollias, et. al.: “MIA-COV19D: COVID-19 Detection through 3-D Chest CT Image Analysis“, 2021
@article{kollias2021mia, title={MIA-COV19D: COVID-19 Detection through 3-D Chest CT Image Analysis}, author={Kollias, Dimitrios and Arsenos, Anastasios and Soukissian, Levon and Kollias, Stefanos}, journal={arXiv preprint arXiv:2106.07524}, year={2021}}
- D. Kollias, et. al.: “Deep transparent prediction through latent representation analysis”, 2020
@article{kollias2020deep, title={Deep transparent prediction through latent representation analysis}, author={Kollias, Dimitrios and Bouas, N and Vlaxos, Y and Brillakis, V and Seferis, M and Kollia, Ilianna and Sukissian, Levon and Wingate, James and Kollias, S}, journal={arXiv preprint arXiv:2009.07044}, year={2020}}
- D. Kollias, et. al.: “Transparent Adaptation in Deep Medical Image Diagnosis”, 2020
@inproceedings{kollias2020transparent, title={Transparent Adaptation in Deep Medical Image Diagnosis.}, author={Kollias, Dimitris and Vlaxos, Y and Seferis, M and Kollia, Ilianna and Sukissian, Levon and Wingate, James and Kollias, Stefanos D}, booktitle={TAILOR}, pages={251–267}, year={2020}}
- D. Kollias, et. al.: “Deep neural architectures for prediction in healthcare”, 2018
@article{kollias2018deep, title={Deep neural architectures for prediction in healthcare}, author={Kollias, Dimitrios and Tagaris, Athanasios and Stafylopatis, Andreas and Kollias, Stefanos and Tagaris, Georgios}, journal={Complex \& Intelligent Systems}, volume={4}, number={2}, pages={119–131}, year={2018}, publisher={Springer}}